Tips and Tricks
There is more to ‘pandas.read_csv()’ than meets the eye
A deep dive into some of the parameters of the read_csv function in pandas

Pandas is one of the most widely used libraries in the Data Science ecosystem. This versatile library gives us tools to read, explore and manipulate data in Python. The primary tool used for data import in pandas is read_csv().This function accepts the file path of a comma-separated value, a.k.a, CSV file as input, and directly returns a panda’s dataframe. A comma-separated values (CSV) file is a delimited text file that uses a comma to separate values.
The pandas.read_csv()has about 50 optional calling parameters permitting very fine-tuned data import. This article will touch upon some of the lesser-known parameters and their usage in data analysis tasks.
pandas.read_csv() parameters
The syntax for importing a CSV file in pandas using default parameters is as follows:
import pandas as pd
df = pd.read_csv(filepath)1. verbose
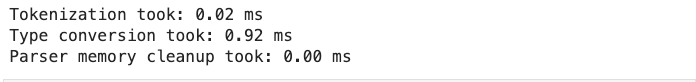
The verbose parameter, when set to True prints additional information on reading a CSV file like time taken for:
- type conversion,
- memory cleanup, and
- tokenization.
import pandas as pd
df = pd.read_csv('fruits.csv',verbose=True)
2. Prefix
The header is a row in a CSV file containing information about the contents in every column. As the name suggests, it appears at the top of the file.
Sometimes a dataset doesn’t contain a header. To read such files, we have to set the header parameter to none explicitly; else, the first row will be considered the header.
df = pd.read_csv('fruits.csv',header=none)
df

The resulting dataframe consists of column numbers in place of column names, starting from zero. Alternatively, we can use the prefix parameter to generate a prefix to be added to the column numbers.
df = pd.read_csv('fruits.csv',header=None, prefix = 'Column')
df
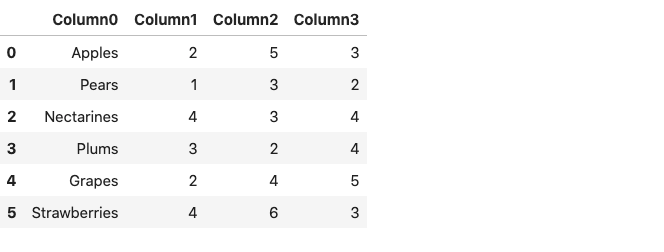
Note that instead of Column, you can specify any name of your choice.
3. mangle_dupe_cols
If a dataframe consists of duplicate column names — ‘X’,’ X’ etcmangle_dupe_cols automatically changes the name to ‘X’, ‘X1’ and differentiate between the repeated columns.
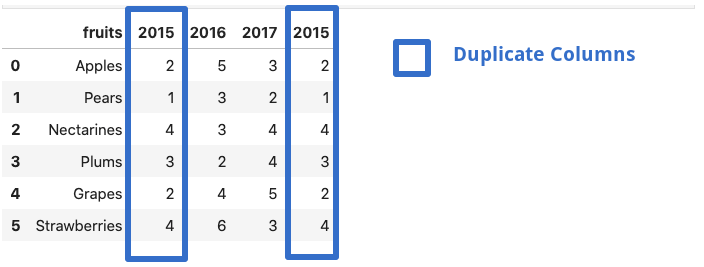
df = pd.read_csv('file.csv',mangle_dupe_cols=True)
df
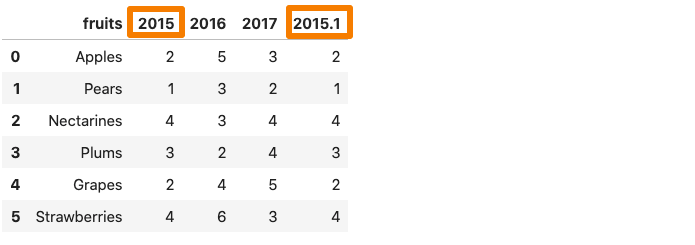
One of the 2015 column in the dataframe get renames as 2015.1.
4. chunksize
The pandas.read_csv() function comes with a chunksize parameter that controls the size of the chunk. It is helpful in loading out of memory datasets in pandas. To enable chunking, we need to declare the size of the chunk in the beginning. This returns an object we can iterate over.
chunk_size=5000
batch_no=1
for chunk in pd.read_csv('yellow_tripdata_2016-02.csv',chunksize=chunk_size):
chunk.to_csv('chunk'+str(batch_no)+'.csv',index=False)
batch_no+=1
In the example above, we choose a chunk size of 5000, which means at a time, only 5000 rows of data will be imported. We obtain multiple chunks of 5000 rows of data each, and each chunk can easily be loaded as a pandas dataframe.
df1 = pd.read_csv('chunk1.csv')
df1.head()
You can read more about chunking in the article mentioned below:
5. compression
A lot of times, we receive compressed files. Well, pandas.read_csv can handle these compressed files easily without the need to uncompress them. The compression parameter by default is set to infer, which can automatically infer the kind of files i.e gzip , zip , bz2 , xz from the file extension.
df = pd.read_csv('sample.zip') or the long form:df = pd.read_csv('sample.zip', compression='zip')
6. thousands
Whenever a column in the dataset contains a thousand separator, pandas.read_csv() reads it as a string rather than an integer. For instance, consider the dataset below where the sales column contains a comma separator.
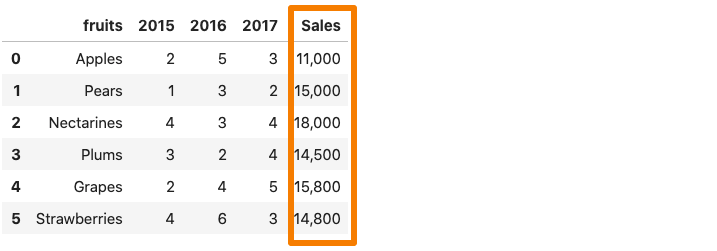
Now, if we were to read the above dataset into a pandas dataframe, the Sales column would be considered as a string due to the comma.
df = pd.read_csv('sample.csv')
df.dtypes

To avoid this, we need to explicitly tell the pandas.read_csv() function that comma is a thousand place indicator with the help of the thousands parameter.
df = pd.read_csv('sample.csv',thousands=',')
df.dtypes

7. skip_blank_lines
If blank lines are present in a dataset, they are automatically skipped. If you want the blank lines to be interpreted as NaN, set the skip_blank_lines option to False.
8. Reading multiple CSV files
This is not a parameter but just a helpful tip. To read multiple files using pandas, we generally need separate data frames. For example, in the example below, we call the pd.read_csv() function twice to read two separate files into two distinct data frames.
df1 = pd.read_csv('dataset1.csv')
df2 = pd.read_csv('dataset2.csv')
One way of reading these multiple files together would be by using a loop. We’ll create a list of the file paths and then iterate through the list using a list comprehension, as follows:
filenames = ['dataset1.csv', 'dataset2,csv']
dataframes = [pd.read_csv(f) for f in filenames]
When many file names have a similar pattern, the glob module from the Python standard library comes in handy. We first need to import the glob function from the built-in glob module. We use the pattern NIFTY*.csv to match any strings that start with the prefix NIFTY and end with the suffix .CSV. The ‘*’(asterisk) is a wild card character. It represents any number of standard characters, including zero.
import glob
filenames = glob.glob('NIFTY*.csv')
filenames
--------------------------------------------------------------------
['NIFTY PHARMA.csv',
'NIFTY IT.csv',
'NIFTY BANK.csv',
'NIFTY_data_2020.csv',
'NIFTY FMCG.csv']
The code above makes it possible to select all CSV filenames beginning with NIFTY. Now, they all can be read at once using the list comprehension or a loop.
dataframes = [pd.read_csv(f) for f in filenames]
Conclusion
In this article, we looked at a few parameters of the pandas.read_csv() function. it is a beneficial function and comes with a lot of inbuilt parameters which we seldom use. One of the primary reasons for not doing so is because we rarely care to read the documentation. It is a great idea to explore the documentation in detail to unearth the vital information that it may contain.
